ggseg.formats provides the ggseg_atlas S3 class that powers the ggseg ecosystem for 2D and 3D brain visualisation. It ships three bundled atlases, a set of accessor functions for querying atlas contents, and a pipe-friendly manipulation API for subsetting, renaming, and enriching atlas objects.
Bundled atlases
The package includes three atlases covering the main atlas types:
- dk — Desikan-Killiany cortical parcellation (68 regions)
- aseg — FreeSurfer automatic subcortical segmentation
- tracula — TRACULA white matter tract atlas
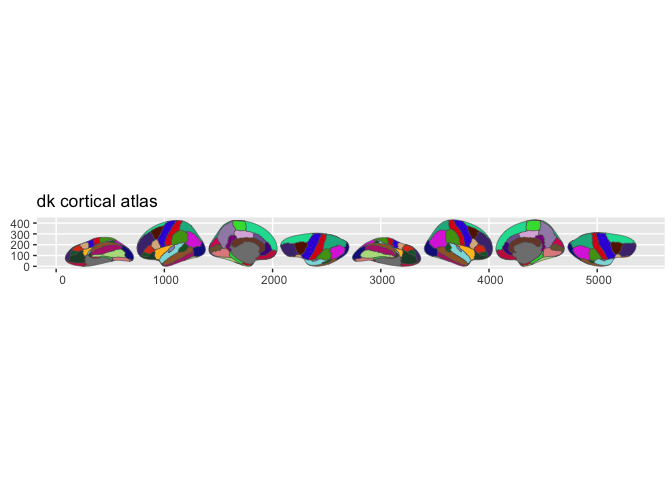
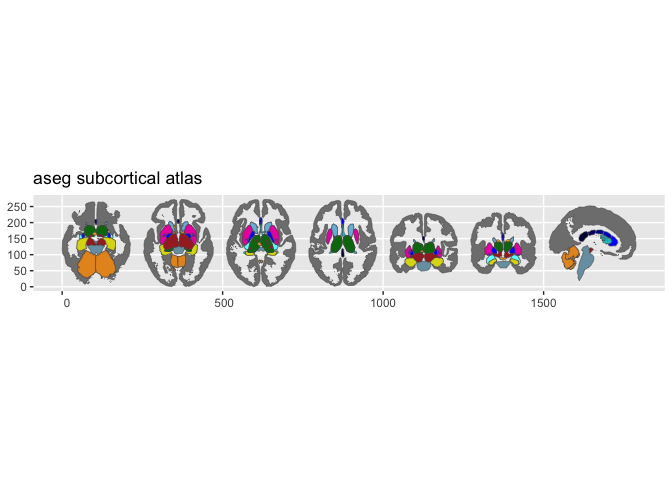
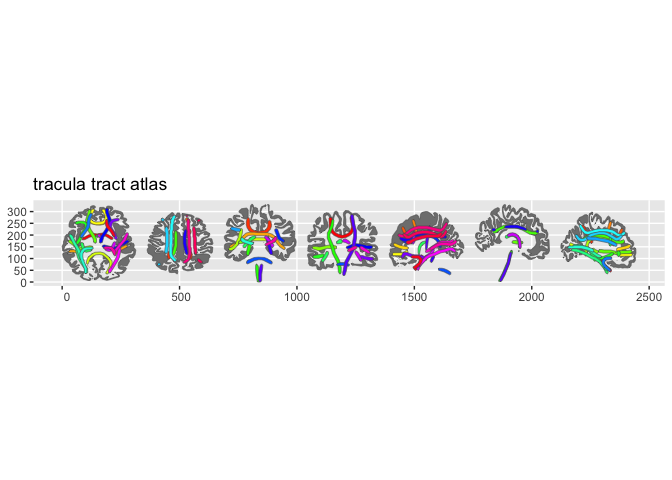
Quick example
Atlas objects are designed for exploration and customisation. You can query regions, filter views, and pipe operations together:
aseg_small <- aseg() |>
atlas_region_keep("hippocampus|amygdala|thalamus") |>
atlas_view_keep("coronal_3|axial_3") |>
atlas_view_gather()
plot(aseg_small)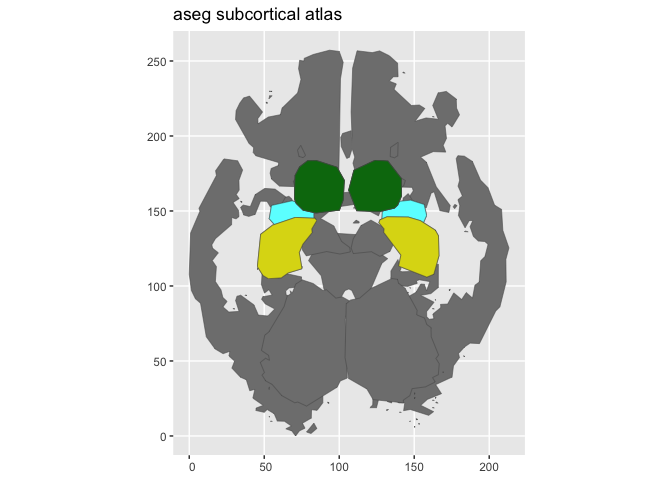
Installation
Install from the ggsegverse r-universe:
options(repos = c(
ggsegverse = 'https://ggsegverse.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
install.packages('ggseg.formats')Or install the development version from GitHub:
# install.packages("devtools")
devtools::install_github("ggsegverse/ggseg.formats")Learn more
-
vignette("ggseg.formats")— understanding atlas structure and accessors -
vignette("atlas-manipulation")— region, view, and metadata manipulation